Please cite:
Schudoma et al.,
Nucl. Acids Res. 38: 970-980.
DOI 10.1093/nar/gkp1010.
Schudoma et al.,
Nucl. Acids Res. 38: 970-980.
DOI 10.1093/nar/gkp1010.
| '6-4-Internal Loop pdb3d5a1A.n369-376 | |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source: [PDB-id:chain] | 3d5a:A (&rarr PDB) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Source: Information | STRUCTURAL BASIS FOR TRANSLATION TERMINATION ON THE 70S RIBOSOME. THIS FILE CONTAINS THE 30S SUBUNIT, RELEASE FACTOR 1 (RF1), TWO TRNA, AND MRNA MOLECULES OF ONE 70S RIBOSOME. THE ENTIRE CRYSTAL STRUCTURE CONTAINS TWO 70S RIBOSOMES AS DESCRIBED IN REMARK 400. | ||||||||||||||||||||||||||||||||||||||||||||||||
| Source: Compound | 16S RRNA | ||||||||||||||||||||||||||||||||||||||||||||||||
| Source: Resolution | 3.21 ANGSTROMS. | ||||||||||||||||||||||||||||||||||||||||||||||||
| Position | (362, 381), (369, 376) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Primary structure ('_': anchors) | _CAAUGG_-_CUGA_ | ||||||||||||||||||||||||||||||||||||||||||||||||
|
Bases with unusual sugar puckers (Standard: C3'-endo) |
2: C2'-endo, 12: C3'-exo | ||||||||||||||||||||||||||||||||||||||||||||||||
|
Bases with unusual glycosidic-bond configuration (Standard: anti) |
12: syn | ||||||||||||||||||||||||||||||||||||||||||||||||
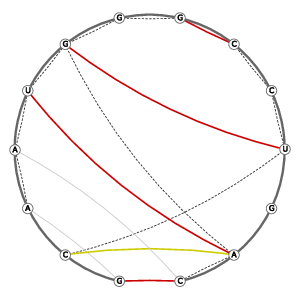
| Tertiary structure: Stacked bases |
|
||||||||||||||||||||||||||||||||||||||||||||||||
|
Tertiary structure: Base-pairs (anchor pairs) |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| Downloads |
Atom coordinates (PDB format) Contact annotation (MC-Annotate format) |
||||||||||||||||||||||||||||||||||||||||||||||||
| 3D Structure | Structure Graph | ||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||
| Structural Clusters | |||||||||||||||||||||||||||||||||||||||||||||||||


