Please cite:
Schudoma et al.,
Nucl. Acids Res. 38: 970-980.
DOI 10.1093/nar/gkp1010.
Schudoma et al.,
Nucl. Acids Res. 38: 970-980.
DOI 10.1093/nar/gkp1010.
| '12-Hairpin pdb2qbh1A.e316-329 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source: [PDB-id:chain] | 2qbh:A (&rarr PDB) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Source: Information | CRYSTAL STRUCTURE OF THE BACTERIAL RIBOSOME FROM ESCHERICHIA COLI IN COMPLEX WITH GENTAMICIN AND RIBOSOME RECYCLING FACTOR (RRF). THIS FILE CONTAINS THE 30S SUBUNIT OF THE FIRST 70S RIBOSOME, WITH GENTAMICIN BOUND. THE ENTIRE CRYSTAL STRUCTURE CONTAINS TWO 70S RIBOSOMES AND IS DESCRIBED IN REMARK 400. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Source: Compound | 16S RRNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Source: Resolution | 4.00 ANGSTROMS. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Position | (316, 329) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Primary structure ('_': anchors) | _ACUGAGACACGG_ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Bases with unusual sugar puckers (Standard: C3'-endo) |
8: C2'-endo, 9: C3'-exo, 10: C3'-exo | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Bases with unusual glycosidic-bond configuration (Standard: anti) |
8: syn, 9: syn | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tertiary structure: Stacked bases |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Tertiary structure: Base-pairs (anchor pairs) |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Downloads |
Atom coordinates (PDB format) Contact annotation (MC-Annotate format) |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3D Structure | Structure Graph | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
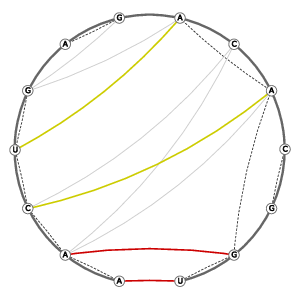
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structural Clusters | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||


