Please cite:
Schudoma et al.,
Nucl. Acids Res. 38: 970-980.
DOI 10.1093/nar/gkp1010.
Schudoma et al.,
Nucl. Acids Res. 38: 970-980.
DOI 10.1093/nar/gkp1010.
| '4-4-Internal Loop pdb2pxt1B.n15-38 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source: [PDB-id:chain] | 2pxt:B (&rarr PDB) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Source: Information | VARIANT 15 OF RIBONUCLEOPROTEIN CORE OF THE E. COLI SIGNAL RECOGNITION PARTICLE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Source: Compound | 4.5 S RNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Source: Resolution | 2.50 ANGSTROMS. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Position | (17, 36), (22, 31) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Primary structure ('_': anchors) | _CAGG_-_AGCA_ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Bases with unusual sugar puckers (Standard: C3'-endo) |
None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Bases with unusual glycosidic-bond configuration (Standard: anti) |
None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tertiary structure: Stacked bases |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Tertiary structure: Base-pairs (anchor pairs) |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Downloads |
Atom coordinates (PDB format) Contact annotation (MC-Annotate format) |
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3D Structure | Structure Graph | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
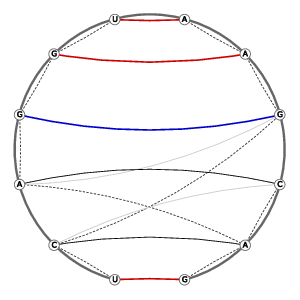
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structural Clusters | |||||||||||||||||||||||||||||||||||||||||||||||||||||||


