Please cite:
Schudoma et al.,
Nucl. Acids Res. 38: 970-980.
DOI 10.1093/nar/gkp1010.
Schudoma et al.,
Nucl. Acids Res. 38: 970-980.
DOI 10.1093/nar/gkp1010.
| '6-8-11-7-4-Multiloop pdb1vq710.m2059-2510 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source: [PDB-id:chain] | 1vq7:0 (&rarr PDB) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Source: Information | THE STRUCTURE OF THE TRANSITION STATE ANALOGUE "DCA" BOUND TO THE LARGE RIBOSOMAL SUBUNIT OF HALOARCULA MARISMORTUI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Source: Compound | 23S RIBOSOMAL RNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Source: Resolution | 2.50 ANGSTROMS. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Position | (2060, 2509), (2067, 2344), (2353, 2394), (2406, 2478), (2486, 2504) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Primary structure ('_': anchors) | _GAAGAC_-_GAUAACAG_-_ACCUCGAUGUC_-_GGGUUUA_-_GGCU_ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Bases with unusual sugar puckers (Standard: C3'-endo) |
4: C2'-endo, 5: C1'-exo, 10: C2'-endo, 11: C2'-endo, 12: C2'-endo, 20: C2'-endo, 21: C2'-exo, 24: O4'-endo, 25: C4'-exo, 26: C2'-endo, 27: C2'-endo, 33: C2'-endo, 37: C2'-endo, 44: C2'-endo, 45: C2'-endo | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Bases with unusual glycosidic-bond configuration (Standard: anti) |
26: syn, 33: syn, 44: syn | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tertiary structure: Stacked bases |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
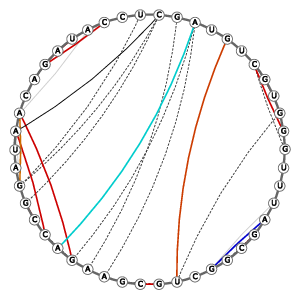
Tertiary structure: Base-pairs (anchor pairs) |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Downloads |
Atom coordinates (PDB format) Contact annotation (MC-Annotate format) |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3D Structure | Structure Graph | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structural Clusters | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||


